Activities
New publishment in Cell Research 在Cell Research新发表文章一篇

Link to this article:
https://doi.org/10.1038/s41422-022-00676-0
Chromatin in the eukaryotic nucleus is spatially organized into three-dimensional (3D) structures including compartments (A or B), topologically associating domains (TADs), and loops.1-3 These structures affect gene expression and are frequently disrupted in developmental disorders and cancer. 4-6 The compartment can be divided into A and B compartment, respectively. The A compartment is transcriptionally active, while the B compartment is transcriptionally silent. A compartments and B compartments interact with other A and B compartments, respectively. However, there are relatively fewer interactions between the A and B compartments.1 Cell differentiation is accompanied by changes in long-range chromatin interactions between compartments. “Loop extrusion” is a well-known model to demonstrate the organization mechanism of chromatin structures, but it merely explains the formation of the short-range interactions, including loops and TADs.3,7-10 Nevertheless, the mechanisms underlying the organization of general long-range chromatin interactions between compartments are not well characterized.
CTCF is a vital structural factor in the canonical “loop extrusion” model, and it organizes chromatin looping via mediating cohesin retention on DNA.7,11,12 On 29 June 2022, Junjun Ding group from Sun Yat-sen University and Cheng Li group from Peking University, published a paper in Cell Research entitled “CTCF organizes Inter-A Interactions Through RYBP-Dependent Phase Separation”. For the first time, they found that, in addition to organizing loops, CTCF is also able to organize long-range chromatin interactions between A compartments via phase separation behavior.
In this study, they generated high-quality CTCF HiChIP data, and found that CTCF organizes inter-A compartment interactions. Then, they analyzed Hi-C data, and found that CTCF depletion reproducibly decreases the interactions between A compartments. However, the interactions between A compartments participated by CTCF can’t be explained by the loop extrusion model, indicating that CTCF may mediate the interactions between A compartments independent of the loop extrusion model.
To explore the mechanism of CTCF-mediated long-range chromatin interactions, they found that CTCF exhibits phase separation behavior in the nucleus, but the purified CTCF in vitro is difficult to form droplets, which raised the possibility that additional factors facilitate the phase separation of CTCF in vivo. Based on the analysis of CTCF partners, they found that RYBP exhibits the highest percentage of intrinsically disordered amino acid residues among all candidate proteins, as well as the phase separation behavior both in vivo and in vitro. They further proved that RYBP facilitates CTCF to undergo phase separation. Furthermore, Hi-C and CTCF HiChIP were performed in RYBP-depleted ESCs, which revealed that CTCF-mediated inter-A compartment interactions also depend on RYBP. To explore the role of CTCF phase separation in the formation of interactions between A compartment, they disrupted CTCF phase separation via RYBP depletion to attenuate inter-A compartment interactions. Subsequently, they exogenously expressed a fusion protein of CTCF with HNRNPA1 IDR to establish an artificially induced CTCF phase separation system in endogenous RYBP-depleted ESCs. Induced CTCF phase separation restores inter-A compartment interactions impaired by RYBP depletion. Upon the further disruption of phase separation in ESCs with induced CTCF phase separation, the interactions between A compartments reduced again. Thereby, it supports the model that CTCF organizes inter-A compartment interactions via phase separation.
They further demonstrated that CTCF phase separation is involved in the regulation of embryonic stem cell (ESC) pluripotency. CTCF phase separation is seemingly abundant in ESCs, and decreased in differentiated cells. In ESCs, disrupting CTCF phase separation via RYBP deficiency down-regulates a majority of genes that are adjacent to the anchors of inter-A compartment interactions, and the down-regulated genes are enriched in cell proliferation terms. Disruption of CTCF phase separation significantly impaired the proliferation ability of ESCs, while induction of CTCF phase separation in RYBP-deficient cells restored their self-renewal ability. And induced CTCF phase separation inhibits the differentiation of ESCs to NPCs. Therefore, CTCF phase separation maintains embryonic stem cell self-renewal, and inhibits their differentiation to neural progenitor cells.
Then they further analyzed the relationship between RYBP-dependent CTCF phase separation and Polycomb body. RYBP was initially identified as a component of the Polycomb complex, which represses the expression of genes involved in neuronal development.13,14 And the components of the Polycomb complex can be aggregated in Polycomb body to form phase separation. Thus they explored whether the functions of RYBP in CTCF phase separation are still restricted in Polycomb body. They found that RYBP-CTCF co-binding peaks at chromatin showed low enrichment of the core modifying enzyme in the Polycomb complex RING1B, but these sites are enriched with transcriptional activation-associated histone modifications. The majority of the RYBP-CTCF co-localized puncta lacked RING1B in the nucleus, and there are obvious differences in the expression of altered genes between that are from disrupting CTCF phase separation and disturbing Polycomb complex. Therefore, functions of RYBP in CTCF phase separation are primarily different from Polycomb body.
Altogether, the present work initially demonstrated the new function of CTCF in organizing long-range chromatin interactions between active chromatin compartments, and raised a novel model that CTCF organizes inter-A compartment interactions via RYBP-mediated phase separation. It also revealed that manipulating CTCF phase separation could regulate the pluripotency of embryonic stem cells. Thereby, at the level of the compartments, they fill the theoretical blank for the mechanism of 3D chromatin structure formation.
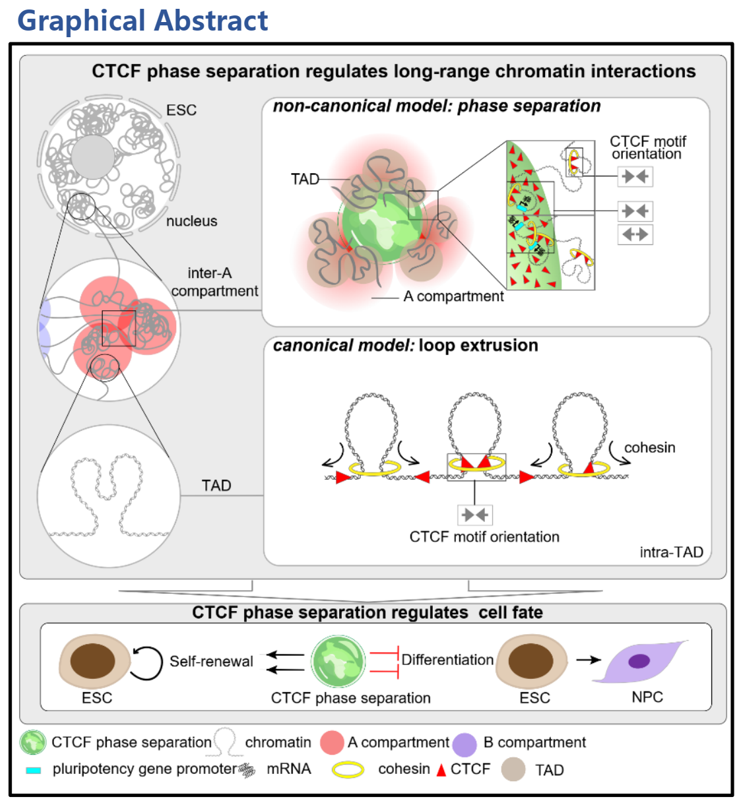
真核细胞中的染色体通常会在空间上折叠成有序的三维结构,包括区室(compartment)、拓扑相关结构域(Topological-Associated Domain, TAD)及染色质环(chromatin loop)等1-3。这些结构广泛参与基因的表达调控,而结构异常则会导致发育异常甚至是肿瘤的发生4-6。对于compartment结构,又可细分为A与B 两类。其中A compartment中基因表达相对活跃,而B compartment中基因表达则相对沉默。结构上A compartment倾向于A compartment之间形成互作,B compartment倾向与B compartment形成互作,而 A与B compartment之间的互作则相对较少1。Compartment之间的互作在细胞命运转变过程中也会发生动态的变化。“环挤压”模型是目前被广泛认可的解释染色质是如何折叠的经典模型,但其仅可解释染色质环及拓扑相关结构域这些相对短距离的染色质之间互作的形成过程3,7-10。对于Compartment之间这种长距离的染色质互作是通过什么因子以及通过什么机制形成的,仍缺乏解释。
CTCF是经典的“环挤压”模型中的重要结构因子,参与介导cohesin在DNA上的驻留从而形成染色质环7,11,12。2022年6月29日,中山大学的丁俊军课题组与北京大学的李程课题组在Cell Research上发表了题为 CTCF organizes inter-A compartment interactions through RYBP-dependent phase separation的文章,首次发现CTCF除了介导经典的环挤压模型调控短距离染色质互作,还可以通过非经典的相分离模型介导长距离的A compartment之间的互作。
在该项研究中,研究人员首先分析了CTCF的HiChIP数据,发现CTCF广泛参与A compartment之间的长距离互作。进一步分析Hi-C数据,结果表明CTCF敲除后会减弱A compartment之间的互作。但是CTCF参与的A compartment之间的互作不符合经典的“环挤压”模型,预示着CTCF可能以一种独立于“环挤压”模型的方式介导A compartment之间的互作。
为探究CTCF介导长距离互作的具体机制,研究者发现CTCF在细胞核内表现出相分离的特点,但体外纯化的蛋白很难形成液滴。预示可能存在其它因子辅助CTCF相分离。通过分析CTCF的互作蛋白,发现其中的RYBP具有最高比例的无序氨基酸,且在体内外均表现出相分离的特点,并进一步证明了CTCF的相分离依赖于RYBP。此外,通过分析RYBP敲除后的Hi-C及CTCF的HiChIP数据,发现CTCF介导的A compartment之间的互作也依赖于RYBP。为进一步探究CTCF的相分离在A compartment间互作形成中的作用,研究者首先通过敲除RYBP的方式破坏CTCF的相分离以减弱A compartment之间的互作。接下来在RYBP敲除细胞中融合表达CTCF与HNRNPA1 IDR以构建人工诱导CTCF相分离系统,发现诱导CTCF相分离可显著恢复A compartment之间互作。而利用1,6-hex进一步破坏造相后细胞中的相分离,A compartment之间的互作又重新减弱。从而证明了CTCF通过相分离的方式调控A compartment之间的互作。
研究者进一步证实了CTCF相分离可参与干细胞多能性调控。CTCF相分离倾向于在胚胎干细胞中富集,而在神经祖细胞中CTCF相分离能力则明显减弱。在胚胎干细胞中,通过敲除RYBP破坏CTCF相分离,大量临近A compartment之间互作anchor的基因下调,且下调的基因富集在细胞增殖相关通路上。增殖实验结果表明,破坏CTCF相分离可显著降低胚胎干细胞的自我更新能力,而诱导CTCF相分离则显著恢复胚胎干细胞的自我更新能力。在胚胎干细胞向神经祖细胞分化的过程中,诱导CTCF相分离则可抑制其分化。因此,CTCF相分离维持干细胞的自我更新,并抑制其向神经祖细胞分化。
接下来研究者进一步解析了RYBP依赖的CTCF相分离与Polycomb body的关系。RYBP一直被认为是Polycomb 复合物的成分,抑制发育基因的表达13,14。而Polycomb 复合物的成分可聚集在Polycomb body中形成相分离,因此CTCF相分离本质上是否就是Polycomb body?研究者发现RYBP与CTCF在染色质上共富集的位点低富集Polycomb 复合物中的核心修饰酶RING1B,但高富集转录激活的组蛋白修饰;大部分RYBP与CTCF共聚集的puncta缺乏RING1B;干扰CTCF相分离与干扰Polycomb 复合物后的差异表达基因存在很大的差异。因此证明RYBP依赖的CTCF相分离与Polycomb body存在着明显的差异。
综上所述,该研究首次揭示了CTCF具有调控compartment之间长距离互作的新功能,并提出了CTCF通过RYBP依赖的相分离的方式调控染色质长距离互作的新模型。同时揭示了操控CTCF的相分离可调控胚胎干细胞的多能性。从而在compartment层级上,填补了染色质三维结构是如何形成的重要理论空白。
References:
1 Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289-293, doi:10.1126/science.1181369 (2009).
2 Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376-380, doi:10.1038/nature11082 (2012).
3 Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665-1680, doi:10.1016/j.cell.2014.11.021 (2014).
4 Weintraub, A. S. et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171, 1573-+, doi:10.1016/j.cell.2017.11.008 (2017).
5 Lupianez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012-1025, doi:10.1016/j.cell.2015.04.004 (2015).
6 Flavahan, W. A. et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110-+, doi:10.1038/nature16490 (2016).
7 Rao, S. S. P. et al. Cohesin Loss Eliminates All Loop Domains. Cell 171, 305-+, doi:10.1016/j.cell.2017.09.026 (2017).
8 Fudenberg, G. et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep 15, 2038-2049, doi:10.1016/j.celrep.2016.04.085 (2016).
9 Kim, Y., Shi, Z., Zhang, H., Finkelstein, I. J. & Yu, H. Human cohesin compacts DNA by loop extrusion. Science 366, 1345-1349, doi:10.1126/science.aaz4475 (2019).
10 Davidson, I. F. et al. DNA loop extrusion by human cohesin. Science 366, 1338-1345, doi:10.1126/science.aaz3418 (2019).
11 Tang, Z. H. et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell 163, 1611-1627, doi:10.1016/j.cell.2015.11.024 (2015).
12 Lomvardas, S. et al. Interchromosomal interactions and olfactory receptor choice. Cell 126, 403-413, doi:10.1016/j.cell.2006.06.035 (2006).
13 Neira, J. L. et al. The Transcriptional Repressor RYBP Is a Natively Unfolded Protein Which Folds upon Binding to DNA. Biochemistry-Us 48, 1348-1360, doi:10.1021/bi801933c (2009).
14 Gao, Z. et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45, 344-356, doi:10.1016/j.molcel.2012.01.002 (2012).
